
Hunting for Antibiotics in the World’s Dirtiest Places
“Compost bin. Pig trough. Dog-food bowl. Laptop keyboard.”
On a chilly autumn morning in northwest London, just outside the Euston train station, Adam Roberts stops at the top of an outdoor staircase, looks around for police, and tries to appear inconspicuous. This is harder than it sounds, and not only because he’s 6 foot 3.
Roberts pulls a plastic-wrapped package from his pocket, tears it open, and slides out a long, slender tube and a swab that looks like an overgrown Q-tip. After checking again for anyone watching, he trots down the stairs, dragging the swab along the handrail, and slips the swab into the tube and the tube into his pocket. Then he strolls away.
After a block, Roberts veers off busy Euston Road and down side streets toward his lab at University College London. He’s not up to anything nefarious—quite the opposite—but with closed-circuit TVs everywhere and London on high alert for terrorist threats, he worries about raising alarms. Only once he reaches the middle of St. George’s Gardens, a green space of big trees and old tombstones, does he relax.
What Roberts has just done, in an action that he and people who support him have performed hundreds of times, is to return to a practice that was abandoned more than 40 years ago. He has sampled the environment, hoping to find in the dirtiest, most germ-filled places an answer to one of the most pressing problems of our day.
Drug resistance—the ability of bacteria to defend themselves against the compounds we use to kill them—has impaired the effectiveness of almost every antibiotic produced since the first ones were developed, in the 1940s. At least 700,000 people are estimated to die worldwide every year from infections that no longer respond to antibiotics. That toll could balloon to more than 10 million a year by 2050 if we can’t slow the spread of resistance or find new drugs; routine surgeries and minor injuries will become life-threatening.
Yet making the necessary changes to stave off this catastrophe seems to be beyond us. We continue to take antibiotics with abandon (nearly a third of antibiotic prescriptions in the U.S. aren’t actually needed) and feed huge quantities of them to farm animals. And pharmaceutical companies—daunted by how quickly resistance can undermine drugs that may take a decade and a billion dollars to develop—are not rushing to fill the gap.
That’s where Roberts, a 43-year-old microbiologist from central England, comes in. Back at his lab, he pulls out a handful of tubes that he collected during his walk and labels them: shoe, bathroom-door handle, tree, bench, handrail. He reaches for a stack of petri dishes, each holding a layer of clear-yellow growth medium. One by one, he opens the dishes, swipes the tip of a swab over the agar, closes and marks them, and sets them aside to be incubated.
“The natural microbial world has a whole array of chemistry we haven’t yet looked into,” he tells me, recording the dishes in a ledger. “And we don’t have to go to the bottom of the ocean or an extreme environment to find it.”
There was a time when all of our antibiotics came from natural sources. That is how the antibiotic era began, in 1928: Sir Alexander Fleming reputedly left a window open in his London laboratory, and discovered weeks later that specks of the mold Penicillium, blown in on the breeze, excreted a chemical that killed the bacterium Staphylococcus. That chemical became the first antibiotic, penicillin. It was followed by chloramphenicol, made by a bacterium found in compost in Venezuela, and chlortetracycline, excreted by a bacterium found in a field at the University of Missouri, a test plot for growing hay.
These were the foundational drugs of the antibiotic era, which turned the historical menace of infection from a death sentence into an inconvenience. Their ability to kill disease bacteria was not an accidental property. The earliest antibiotics were refined versions of chemical weapons that bacteria had developed over millennia to fend off other microorganisms as they competed for living space and food. The organisms that made these weapons seemed to thrive in damp and dirty places.
Those early antibiotics were so successful and so lucrative that manufacturers began scouring the world for additional ones. Bristol-Myers (now Bristol-Myers Squibb) included a pre-addressed envelope in its 1951 annual report, along with a note asking shareholders to scoop up “a teaspoon of soil, slightly moist but not wet, and free from large stones,” and mail it to company headquarters. Eli Lilly struck an agreement with members of the Christian and Missionary Alliance: Ministers going to the developing world were given test tubes to take with them. Pfizer recruited explorers, pilots, and foreign correspondents to send it soil samples.
Those searches resulted in antibiotics that remain crucial today: erythromycin, from the Philippines; vancomycin, from the jungle floor in Borneo; daptomycin, from the foothills of Mount Ararat, the biblical resting place of Noah’s Ark. But the work of identifying and isolating useful compounds was punishingly slow. Selman Waksman, a microbiologist who was funded by Merck, processed 10,000 samples before identifying streptomycin. Pfizer went through more than 130,000 samples before finding terramycin, an early tetracycline.
Soil teems with microorganisms—a teaspoon can contain millions—yet only a subset of them make compounds that may prove useful as antibiotics. And within that subset, only a tiny fraction will grow in a lab, away from the complex natural environments in which they evolved. Researchers who thought they were soaring into an entire new universe of cures—and profits—found themselves stuck on the launchpad, discovering the same antibiotic compounds over and over again. The developers of terramycin told The New Yorker in 1951 that before finding it, they had rediscovered streptomycin “at least a hundred times.”
By the end of the 1960s, drug companies gave up on finding antibiotics in soil and turned to assembling synthetic compounds in labs. Not coincidentally, the rate of finding new drugs dropped. From 1940 to 1970, a dozen different classes of antibiotics got all the way through clinical testing and entered the American market. Since 1970, only a few new classes have, which means that almost all the new antibiotics produced since then have been variations on existing ones, with mechanisms that bacteria already have learned to defend against.
With no new drugs to curb their spread, disease bacteria took off. MRSA, multidrug-resistant staph, jumped out of hospitals in the mid-’90s, sidelining athletes and creating cases of fast-moving pneumonia that can kill a child in days. The early 2000s brought the spread of VRE, which causes grave hospital infections and is resistant to vancomycin, a last-resort drug. NDM, a gene that confers resistance to a family of drugs called carbapenems, was transported out of India and around the globe by infected travelers in the late 2000s. Another resistance gene, MCR, which disables the very-last-resort antibiotic colistin (like vancomycin, a holdover from the 1950s), was identified in China in 2015 and has since turned up in more than 30 countries around the world.
By last year, the rising tide of multidrug resistance had caught the attention of the United Nations, which held a rare special summit during a meeting of its General Assembly. The convocation committed world governments to cracking down on antibiotic misuse within their borders and supporting research to find new drugs. Ban Ki-moon, then the secretary-general, called drug resistance “a fundamental, long-term threat to human health.”
“Compost bin. Pig trough. Dog-food bowl. Laptop keyboard.”
Roberts is flipping through a thick binder in his lab, reading off the places where the samples in his collection came from. He didn’t collect most of them himself. They were provided by a network he built, through a crowdsourcing campaign and a Facebook page—a modern version of the drug-company campaigns of the 1950s, aimed at harvesting samples from a wider geographic range than he could ever reach on his own.
Roberts earned his doctorate here at University College London in 2002, and for more than a decade he studied one of the main ways bacteria acquire antibiotic resistance: passing genes back and forth by trading segments of DNA. Transmissible resistance, as it is called, was described in the 1960s by two Japanese researchers, who noticed that strains of the food-borne-illness bacterium Shigella had become resistant to drugs that patients had never received.
It has been a microbiological nightmare ever since. Transmissible resistance allows the mutations that confer protection against antibiotics to spread not just through inheritance, from mother cell to daughter, but also among unrelated bacteria via the exchange of plasmids, small loops of DNA that exist separate from the chromosomes. Plasmids can transport multiple genes at the same time, so they allow resistance against multiple drugs to stack up in bacteria like cards in a winning hand of poker.
Roberts was fascinated by this phenomenon. But after years of study, he decided to shift his focus. “I started to think we could go on forever finding new resistance genes, because they are always evolving,” he says. “Instead of looking for new genes, why not look for new drugs?”
He decided to start where pharmaceutical chemistry had left off decades earlier: in the messy real-world settings where bacteria duke it out. He launched his campaign, called Swab and Send, in February 2015. For £5, participants got a sample tube, a mailing envelope, and an explanation of what Roberts wanted them to look for: a spot in the environment where bacteria were likely to be competing for nutrition and room to reproduce. He asked them to use their imagination. The less sanitary, the better.
In a departure from the first antibiotic searches, Roberts does not ask his sample-collectors to focus on soil. Instead he wants them to search in places his predecessors may have overlooked. “There’s such a rich microbial environment everywhere around us,” he says. “Every single place is a niche, where bacteria will have evolved and adapted independently. Soil may have evolved biological warfare, if you like, completely differently than a marine environment, or a muddy environment, or contaminated pond water. There’s a possibility of different chemistry everywhere.”
The Swab and Send campaign fired people’s enthusiasm: Within two months, Roberts received more than £1,000, and hundreds of swabs. Small checks continue to arrive by mail. (The price of participation has gone up, to £30 for five swabs.) Elementary schools invite Roberts to make presentations, and he gives the kids swabs to take home. He has taken sample tubes to parties and to newsrooms. He has two swabs that were swiped across desks in the Houses of Parliament.
Video: How to Fight Superbugs
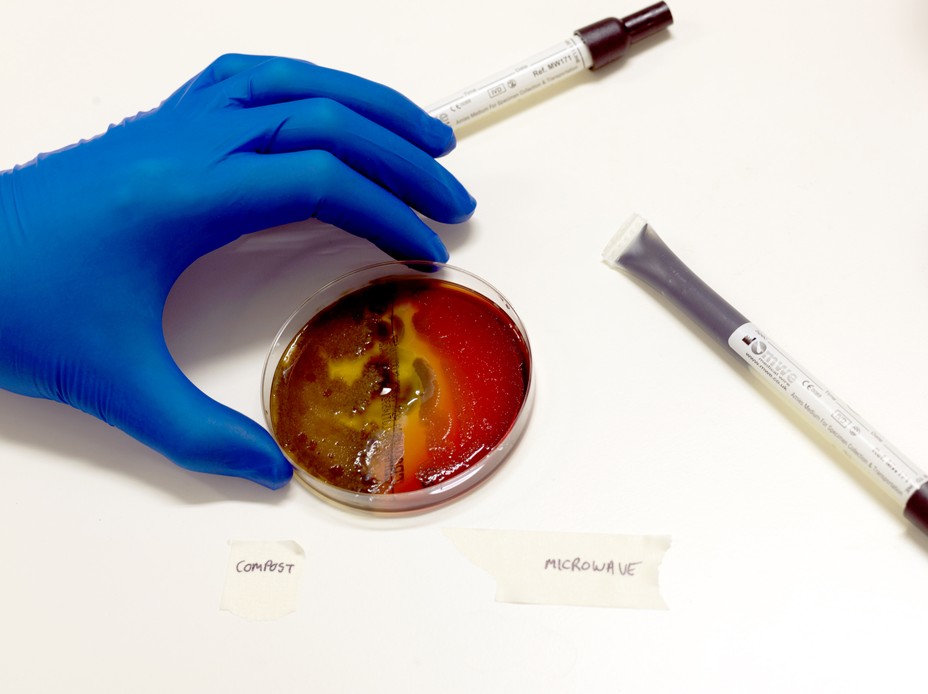
“Toilet in the third tier of the football ground in Manchester,” Roberts reads. (That one arrived with a sketch of a football fan using the toilet.) “Fridge puddle of yuck from a forgotten and moldy lettuce.” We’re sitting in his lab. Roberts is opening petri dishes that have been incubating overnight and transferring samples of whatever bacteria have grown into 96 tiny indentations in a culture plate. He is repeating a process that the scientists of the 1940s followed, growing bacteria in the lab to see what they could do.
The first step is to scribble a swab over a dish of culture medium and let it incubate. The second is to separate all the bacteria that grow on the gel—one swab might have picked up many species—and plop them into individual wells on a fresh plate, to let them multiply without interference from one another. In the third step, Roberts dabs each of these samples into culture dishes that contain another microorganism, to see whether they can hold their own against competition. He’s looking for a “zone of inhibition”: a clear ring around a bacterium indicating that it produces a compound that can kill.
A bacterium that clears that hurdle then faces a higher one: being tested against a strain of E. coli resistant to 15 different drugs. If the bacterium survives that challenge, the compound it produces is considered worthy of further scrutiny. Roberts uses analytic tools that did not exist in the 1940s to find out whether the survivor is really something new.
Since Swab and Send began, Roberts and his graduate students have painstakingly coaxed thousands of bacteria samples through the successive rounds of incubation. Out of all those, hundreds have secreted compounds that killed at least one test bacterium, and a few killed a fungus—potentially precious finds, because antifungal drugs are in even shorter supply than antibiotics. He has found 18 promising bacteria, so far, that killed the multidrug-resistant E. coli.
It is slow work, judged against the pace of bacterial evolution. That frustrates Roberts, because he knows firsthand the risk that resistance poses. Three years ago, his daughter, who was 6 at the time, was playing in the countryside and scratched her leg. The scratch turned into a pustule, and a wash of red began to spread up and down her shin. Doctors tried three different types of antibiotics. None worked. Within 12 hours of her arrival at University College Hospital, she was carted into surgery.
His daughter recovered, though she still has a dent on her shin where the infection was cut out. But for Roberts, the episode brought home how unpredictable antibiotic resistance is, and how short the distance might be between a single recalcitrant infection and the future he’s trying to prevent: multiple tries at treatment, long hospital stays, huge health-care costs, early deaths.
“If the health-care system could not have kept up with her infection, she could have lost her leg,” Roberts says. “The way you get to having a health-care system that can’t keep up is to have it collapse under the pressure of having no antibiotics. I can see that happening really easily.”

The slow pace of Roberts’s research hints at the enormity of the challenge of finding new antibiotics.
“The hard thing isn’t finding things that kill bacteria; steam, fire, bleach can do that,” says John Rex, who led clinical antibiotic development at AstraZeneca and now is the chief medical officer at F2G, a firm working on new antifungal drugs. “The challenge is to find things that kill bacteria but don’t harm the person taking them. You’re talking about a chemical that goes into your mouth, into your gut, into your blood, unchanged, and winds up where the infection is and kills the bacteria, and yet is not toxic to you.”
The beauty of man-made antibiotics was that they could be tailored to those challenges, and to whatever clinical need drug developers perceived. But as antibiotics have fallen to the onslaught of resistance, developers are turning back to natural sources.
“We haven’t come close to tapping the potential of the natural world,” Gerry Wright, the director of the Michael G. DeGroote Institute for Infectious Disease Research at McMaster University in Ontario, told me. Wright is the co-founder of the university’s High Throughput Screening Lab, a lab that can automate the testing of compounds. In the days it takes Roberts and his team to do a few dozen tests, a lab like Wright’s can perform tens of thousands.
“A typical Streptomyces in soil has the genetic programming to produce 20 to 40 different compounds,” Wright says. “Every one of those things has some biological activity. Not all of them are antibiotics, of course, but they’re products of evolution. They’re not kept around for nothing. If you compare that to 30 random chemicals made by a chemist, you’d be lucky if one or two have any biological activity at all.”
These days, Roberts isn’t the only one looking to the natural world for new antibiotics. A team at Northeastern University has invented a device, called the iChip, that allows bacteria that do not thrive in laboratory cultures to be grown in soil; its use led to the isolation of a promising compound, teixobactin, which is still being studied. And a project called the Small World Initiative—created in 2012 by Jo Handelsman, a University of Wisconsin professor who served as the Obama administration’s associate director for science—is similar to Roberts’s campaign, but with more emphasis on education. It teaches basic microbiology to thousands of high-school and college students each year by having them collect soil samples, isolate the bacteria the samples contain, test the bacteria for antibiotic activity, and then present their research at a symposium. The goal is to find novel compounds that companies or academic researchers could take through the next steps.
Even if those efforts succeed, it’s unclear how a new antibiotic would reach the market. The later stages of drug development—the multiphase clinical trials that test the safety and efficacy of a new antibiotic in thousands of patients—require a level of funding that only large pharmaceutical companies can muster, and that they have been reluctant to commit. How best to persuade companies to return to making antibiotics is a hot policy topic in the United States and Europe, with various incentives—grants, prizes, extended patents—all up for debate.
Roberts—who has held Swab and Send together with supporters’ small checks, one grant of £20,000, and stubborn belief—speaks wistfully of the difference that more resources could make for his project. During my visit to London, we sit on high stools in his lab while he holds culture plates up to the ceiling light, looking for telltale zones of inhibition. It’s a motion that Fleming might have made in the 1930s, when antibiotics were too new for anyone to recognize the moneymaking machine they would briefly become.
“If I had a bigger pot of money, it would really improve the amount of work we could do,” Roberts says, pulling another stack of plates toward him. “I would try to create a team with every necessary piece of expertise and all the kit they need. They would all be in one place. They would all be talking to each other. At the moment—which is normal for an academic environment—everything just takes a little bit longer than it should.”
After I return to the U.S., I call Roberts to check in. His mood is utterly different; his voice, and his finances, have gotten a lift. He has been recruited to move to the Liverpool School of Tropical Medicine, to work in a £25 million facility that will enable him to do just what he had envisioned: bring together teams from different scientific disciplines to work on new antibiotics. He is taking Swab and Send with him. The project will have its own budget, and a new lab and new equipment; any promising compounds he finds can be tested and developed by teams already working within the school. (Late-stage trials, with their huge logistical demands, will require a commercial partner.)
When I ask how much these resources might accelerate his search for a new drug, he corrects me.
“We don’t need just one,” he says. “Or five. If we get just a few, we’re still just playing leapfrog. We need a thousand new drugs, so that clinicians can go to the cupboard and say, ‘Right, in this decade, we’re using these 200, and we’re saving the other 800 for when the usefulness of these runs out.’ ”
“I think that is achievable,” he says. “But it’s going to take a massive amount of work.”